A Deep Learning Library for X-Risk Optimization
An open-source library that translates theories to real-world applications
Latest News Install
Why LibAUC?
LibAUC is a novel deep learning library to offer an easier way to directly optimize commonly used performance measures and losses with user-friendly APIs. LibAUC has broad applications in AI for tackling both classic and emerging challenges, such as Classification of Imbalanced Data (CID), Learning to Rank (LTR), and Contrastive Learning of Representation (CLR).
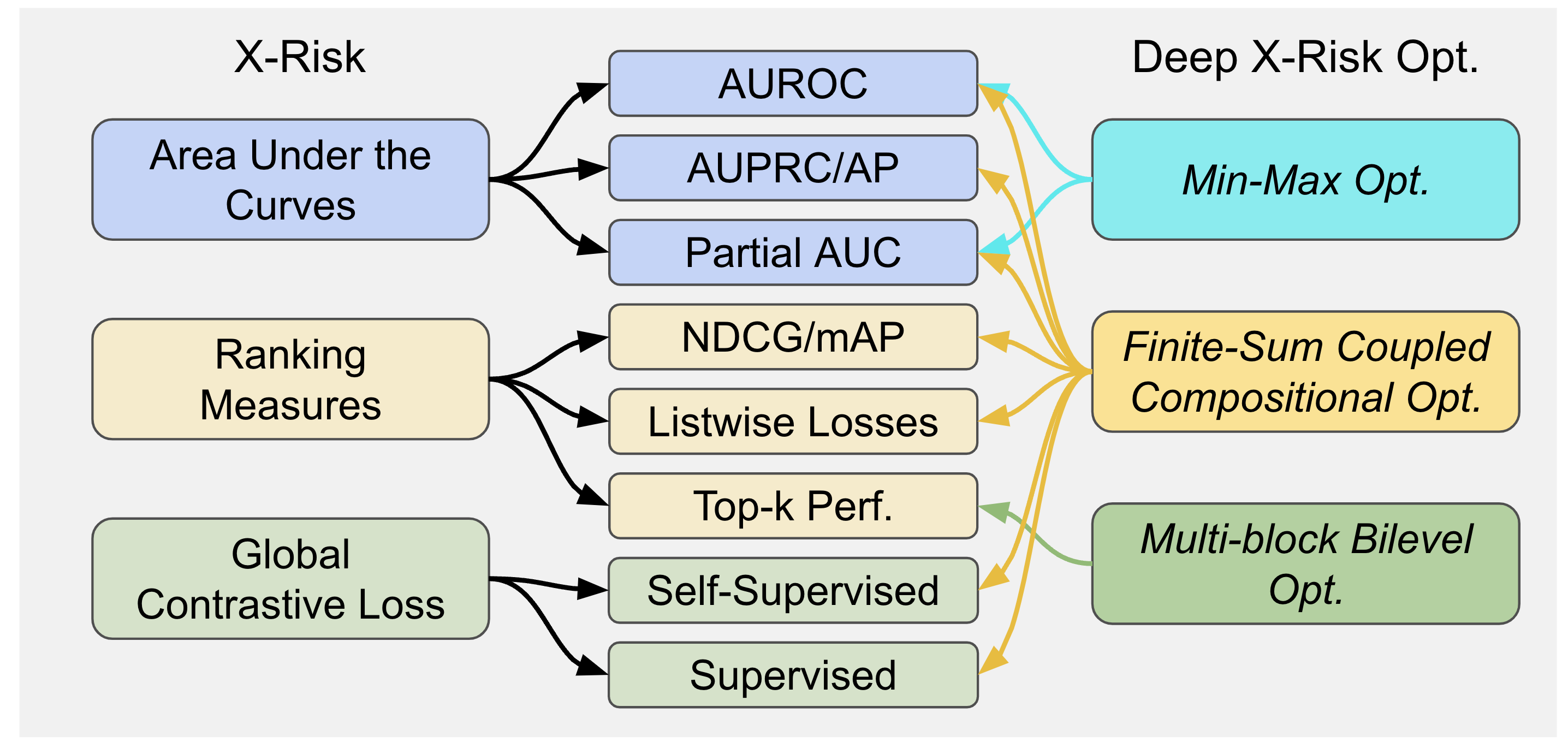
LibAUC provides a unified framework to abstract the optimization of a family of risk functions called X-Risk, including surrogate losses for AUROC, AUPRC/AP, and partial AUROC that are suitable for CID, surrogate losses for NDCG, top-K NDCG, and listwise losses that are used in LTR, and global contrastive losses for CLR. For more details, please check our LibAUC paper.
Key Features
Easy Installation
Easy to install and integrate LibAUC into existing training pipeline using popular Deep Learning frameworks like PyTorch.
Broad Applications
Users can learn different neural network structures (e.g., linear, MLP, CNN, GNN, transformer, etc) that support their data types.
Efficient Algorithms
Stochastic algorithms with provable convergence that support learning with millions of data points without a large batch size.
Hands-on Tutorials
Hands-on tutorials are provided for optimizing a variety of measures and objectives belonging to the family of X-risks.
What is X-Risk?
LibAUC is powered by Empirical X-Risk Minimization (EXM), where X-Risk formally refers to a family of compositional measures in which the loss function of each data point is defined in a way that contrasts the data point with a large number of others. Mathematically, X-Risk optimization can be cast into the following abstract optimization problem:
where is a mapping,
is a simple deterministic function,
denotes a target set of data points, and
denotes a reference set of data points dependent or independent of
.
For mathematical derivations, please check our
EXM paper.

Quick Facts
3+
Challenges winning solution (e.g., Stanford CheXpert, MIT AICures, OGB Graph Property Prediction).
4+
Collaborations and deployments at multiple industrial units, e.g., Google, Uber, Tencent, etc.
25+
Scientific publications on top-tier AI Conferences (e.g., ICML, NeurIPS,ICLR).
98k+
Globally adopted by researchers and developers across 85+ coutries.
Applications
Citations
If any questions, please reach out to Prof. Tianbao Yang. If LibAUC is helpful in your work, please cite the following papers:
@inproceedings{yuan2023libauc, title={LibAUC: A Deep Learning Library for X-risk Optimization}, author={Zhuoning Yuan and Dixian Zhu and Zi-Hao Qiu and Gang Li and Xuanhui Wang and Tianbao Yang}, booktitle={29th SIGKDD Conference on Knowledge Discovery and Data Mining}, year={2023} }
@article{yang2022algorithmic, title={Algorithmic Foundation of Empirical X-risk Minimization}, author={Yang, Tianbao}, journal={Technical Report, arXiv:2206.00439}, year={2022}